Latest News
07|01|25
Phanstiel Lab gets into the brewing business!
As part of a scientific outreach project spearheaded by JP, the lab collaborated with Steel String Brewery on a beer and seltzer, named and labeled by Erika. The beverages can be found at Steel String and stores throughout the Triangle.


06|25|25
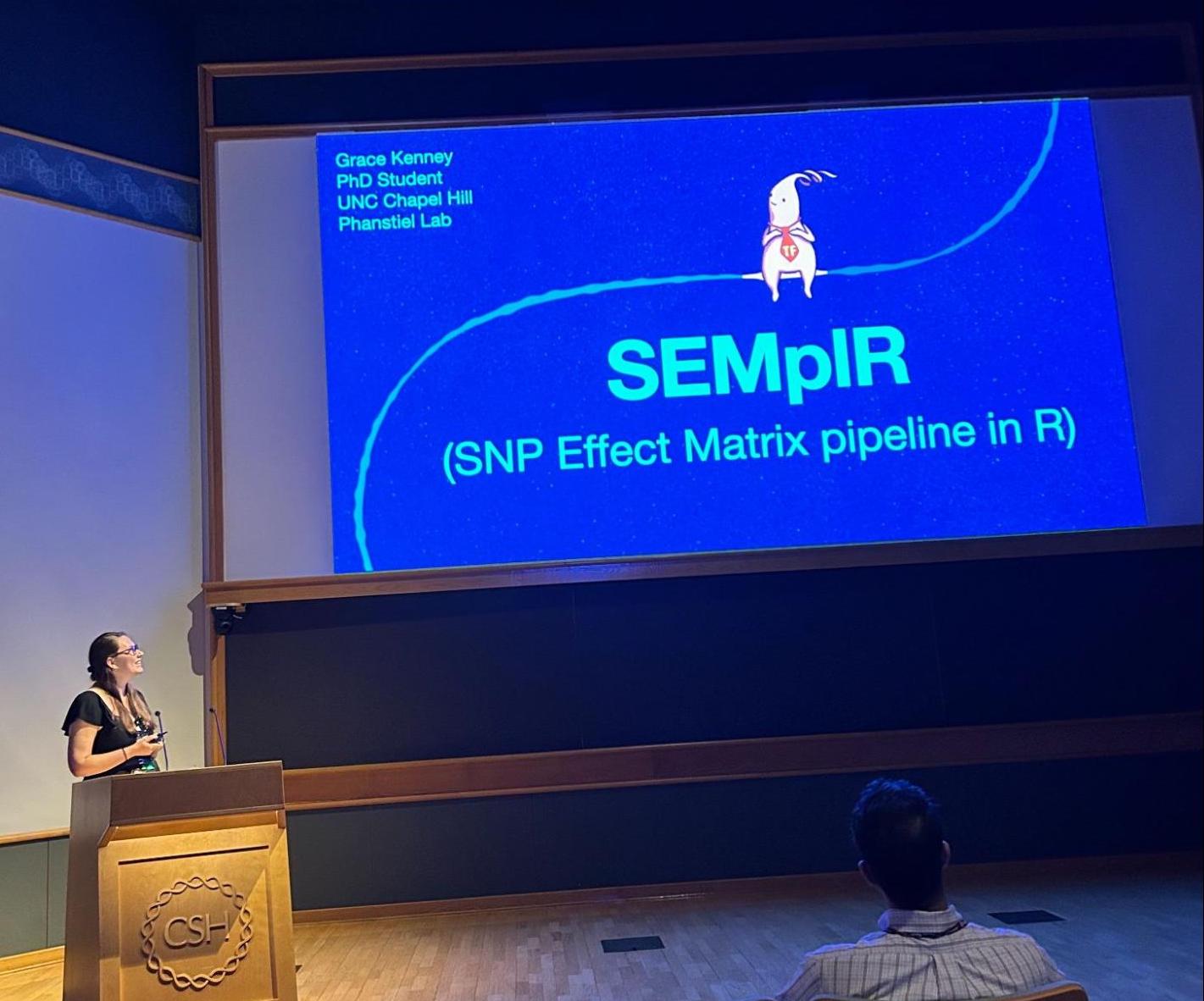
Grace Gives a Talk at Cold Spring Harbor
Grace presented her R bioconductor package SemplR (co-developed with Alan Boyle’s lab at UM) that predicts the impact of genetic variants on transcription factor binding at the Galaxy Bioconductor Community Conference (GBCC) in Cold Spring Harbor
06|02|25
YOSELI ACCEPTED A RESEARCH SCIENTIST POSITION AT UNC SLICE CORE
05|27|25
JESS STARTS A SUMMER INTERNSHIP AT JOHNSON & JOHNSON
Jess is a summer intern on the Biomarker Analytics team within Interventional Oncology, where she works with real world data (RWD) and sequencing data to investigate DNA aneuploidy as a potential biomarker of treatment response in lung cancer.
05|21|25
Gelila Gives a Talk at Annual Prep Symposium
She described the work she did investigating the basis of NUP98-driven AML during her one-year post bacc position in the Phanstiel lab. Great job, Gelila!

03|26|25 – 05|18|25
JP goes on a talk tour across the US
Mar 26 – University Of Alabama at Birmingham
Apr 09 – Brown University
Apr 28 – Washington DC for Aibs Emerging Public Policy Leadership Award (Eppla)
May 18 – University of Utah
03|30|25
ZACK GIVES A TALK AT THE KEYSTONE SYMPOSIA ON INTERPLAY OF CHROMATIN ARCHITECTURE AND TRANSCRIPTION REGULATIONs
Zack was invited to share his work examining the mechanisms of phase-separation driven chromatin loop formation in Banff, AB.
01|08|25
New insights into the genetic basis of osteoarthritis
Our article describing the identification of new putative OA risk genes was published in Cell Genomics.
12|24|24
JP GETS A 2025 CIVIC ENGAGEMENT MICROGRANT FROM RESEARCH!AMERICA
More information found here.
12|11|24
Marielle graduates GMB!
Marielle successfully defended her PhD thesis!
11|08|24
JESS GIVES A PLATFORM TALK AT ASHG
Jess was invited to give a talk at a platform session at the annual American Society for Human Genetics conference!
10|31|24
PHANSTIEL LAB WINS ANNUAL CELL BIOLOGY PUMPKIN CARVING CONTEST
The prize was a catered lunch from the department
10|30|24
TOOBA PASSES BCB CANDIDACY EXAM
09|05|24
JESS GIVES A TALK AT EPIGENETICS OF OSTEOARTHRITIS
Jess was invited to give a talk at the 3rd International Workshop on the Epigenetics of Osteoarthritis in Toronto, Canada

08|14|24
JP's abstract for ASHG is named Reviewer's Choice!
Reviewers scored his abstract in the top 10% of poster abstracts. It will be highlighted in the Exhibit & Poster Hall with a Reviewers’ Choice ribbon.
08|12|24
JP is named an American Society of Human Genetics (ASHG) Human Genetics Scholar!
More information on this can be found here.
08|01|24
JESS RECEIVES AN F31 GRANT FROM NIAMS
07|09|24
JP & Doug are named HHMI Gilliam Fellows!
More information on this can be found here.
07|06|24
Gelila Petros joins the lab as a UNC PREP Scholar
Welcome, Gelila!
06|21|24
JP completes a 6-month internship in the NIH Office of the Director Office of Science Policy
In this role, he helped develop a public vision and framework for including patient and community voices in the design and conduct of NIH-funded clinical research.
06|06|24
Emma, Grace, and Tooba passed their written qualifying exam for BCB!
05|06|24
Chondrocyte response eQTL, chromatin accessibility, and Hi-C preprint is live
Nicole's preprint "Response eQTLs, chromatin accessibility, and 3D chromatin structure in chondrocytes provide mechanistic insight into osteoarthritis risk" went live on bioRxiv! This collaboration with Richard Loeser, Brian Diekman, and many others used genetic and genomic data from primary human chondrocytes in OA-mimicking conditions to interpret OA GWAS variants.
04|15|24
Grace Kenney and Emma Klein offically join the lab!
03|07|24
Tooba wins best poster award at the 7th Annual Carolina Chromatin and Epigenetics Symposium!
08|04|23
Marielle receives an F31 grant from the NIA
Marielle received an F31 grant from the National Institute on Aging for 3 years to study genetic variation in brain cell types in Alzheimer's Disease. Congratulations Marielle!
07|18|23
Welcome Doris Cruz Alonso and Yijia Wu!
Doris is a PREP scholar who just completed her undergraduate studies in Bioengineering at Northeastern University in Boston. Yijia is a research technician studying gene regulation in Alzheimer’s diseases by using iPS-derived brain cell models and high-throughput genomic technologies.


07|12|23
MEGA paper was accepted to Genome Research
Marielle's paper "Chromatin loop dynamics during cellular differentiation are associated with changes to both anchor and internal regulatory features" was published today in Genome Research! This work introduces a deeply sequenced Hi-C time course of megakaryocyte development with matched genomic datasets.


05|26|23
Courtney Grace Neizer and Dakota Watson join for the summer
Courtney-Grace is a visiting summer undergraduate student studying computer science at UNC Charlotte. She is interested in integrating her passion for genetics and computational biology to improve health disparities. She'll be working with JP and Marielle on the visualization and analysis of genomic/non-genomic data. Dakota is a visiting junior biology student at North Carolina A&T State University interested in genome editing. When she's not working, she likes to teach karate classes or watch premier league soccer. She will be working with Zack on gathering imaging data related to phase separation projects.


05|10|23
Tooba Rashid officially joins the lab!
03|13|23
Marielle gives talk at Keystone Symposium
Marielle was invited to give a talk at the Keystone Symposium on Chromatin Architecture in Development and Human Health in Victoria, BC, Canada!
03|02|21
Marielle gives a talk at the Chromatin and Epigenetics Symposium at UNC
Marielle presented her work at the UNC Chromatin and Epigenetics Symposium in a talk titled "A Tale of Two Timecourses"

02|10|23
Nicole and Eric graduate
11|01|22
Paper published in Cell Reports
We published a paper in Cell Reports, "Temporal analysis suggests a reciprocal relationship between 3D chromatin structure and transcription," which describes an eight point time course of changes to 3D chromatin structure during human macrophage activation.
09|20|22
Doug and Dr. Loeser receive NIH NIAMS Grant
Dr. Doug Phanstiel, PI, & Dr. Richard Loeser, Co-PI, received a $2.5 million NIH NIAMS grant: “Identifying novel osteoarthritis risk genes using GWAS, chondrocyte genomics, & genome editing." Their research seeks to identify putative causal knee OA risk variants, map them to their target genes, and quantify their phenotypic impact in chondrocytes. Dr. Brian Diekman and Dr. Karen Mohlke are co-investigators in this project.

09|16|22
Yoseli and JP win best poster awards
Congratulations to Yoseli and JP for their success at the UNC Department of Genetics Retreat!
07|28|22
Nicole wins a Bioconductor award
Nicole accepted a #bioc2022 award at the 2022 Bioconductor Conference for her amazing documentation in plotgardener. Congratulations, Nicole!
07|12|22
Jess wins a spot on the BCB T32 Training Grant
Congratulations Jess!


05|07|22
Katie graduates GMB
Katie Reed is the first student to graduate with a PhD from the Phanstiel lab. Katie got her PhD in Genetics & Molecular Biology, and a certificate in Bioinformatics & Computational Biology. Congratulations Katie!
04|19|22
Jess Byun and JP Flores officially join the lab
Jess joins the lab pursuing a PhD in Bioinformatics & Computational Biology! Jess is from South Korea and graduate from the University of Utah BS in Chemistry and MS in Biomedical informatics. She will be studying multi-omics approaches to understand the molecular mechanisms driving Osteoarthritis. JP joins the lab also pursuing a PhD in Bioinformatics & Computational Biology. He is from Los Angeles, CA and graduated from Occidental College with a BA in Cellular & Molecular Biology in May 2021. He will be studying phase separation-driven chromatin loops and how this contributes to disease. Welcome Jess and JP!
04|08|22
Katie defends her dissertation
Katie presented her dissertation, “Gene Regulation in Time and Space” before her committee, friends and family. Congratulations Dr. Reed!
04|04|22
JP receives an NSF Graduate Research Fellowship
Congratulations to JP Flores, who was awarded an Graduate Research Fellowship from the National Science Foundation! This prestigious award will fund JP for the next three years!
02|22|22
Eric passes BCB candidacy exam
02|04|22
Plotgardener paper published in Bioinformatics
01|12|22
Nicole passes BCB candidacy exam
11|05|21
Katie wins ASHG Predoctoral Trainee Award
Katie won the Charles J. Epstein Trainee Awards for Excellence in Human Genetics Research (predoctoral category) at the ASHG 2021 conference! Among all predoctoral attendees, 30 semifinalists are selected from ASHG conference abstract submissions, and 9 finalists are selected by the Awards Committee to give plenary talks. These presentations are judged at the conference and 3 winners are ultimately selected. Congratulations Katie!
10|20|21
Katie gives plenary talk at ASHG
Katie was invited to give a talk at a plenary session at the annual American Society for Human Genetics conference! Katie was selected for a talk as a Charles J. Epstein Trainee Awards for Excellence in Human Genetics Research finalist. Congratulations Katie!
08|05|21
Nicole gives talk at Bioconductor 2021 Conference
06|23|21
Paper published in Nature
We are excited to share our new paper "Phase separation drives aberrant chromatin looping and cancer development" published today in Nature! The work shows how phase separation of a cancer fusion protein forms chromatin loops that drive oncogenic transcription patterns in severe subtypes of acute myeloid leukemia.

06|11|21
Marielle receives a T32 Training Grant
Marielle Bond re-joins the lab as a graduate student and receives two years of funding from the GM135128 T32 training grant! Marielle will use high throughput genomic techniques to identify the genetic variants contributing to Alzheimer’s Disease.
03|23|21
Sarah receives an NSF Graduate Research Fellowship
Congratulations to Sarah, who was awarded an NSF Graduate Research Fellowship! It will fund her for three years to develop algorithms to predict enhancer-promoter pairs.
01|21|21
Nicole invited to give RStudio talk
05|01|20
Andrea receives UNC Lineberger Cancer Epigenetics Training Grant
Welcome Andrea Perreault, who joins the lab as a postdoc, and congratulations for receiving two years of funding from the UNC Lineberger Cancer Epigenetics Training Grant! She will be studying how phase separation controls 3D chromatin structure and gene expression in cancer.
05|01|20
Welcome Sarah
Sarah Mae Parker joins the lab as a graduate student in the Bioinformatics and Computational biology program. She will be developing tools to improve data visualization and the prediction of enhancer-promoter pairs.
04|15|20
The Lab receives 5 years of funding to study Alzheimer’s Disease
The Phanstiel Lab, in collaboration with the labs of Hyejung Won and Todd Cohen, received 5 years of funding from the National Institute for Aging to study the genetic basis of Alzheimer’s Disease risk. The research applies cutting edge genomic and genome-editing techniques to study transcriptional control in human iPSC-derived microglia responding to Alzheimer’s-related stimuli.
04|15|20
Yoseli receives a two-year postdoctoral fellowship from the BrightFocus Foundation
Yoseli Quiroga will be studying how specific Alzheimer's disease risk and protective variants alter transcriptional regulatory networks in innate immune cells.
03|26|20
Isha receives a SURF fellowship
Isha Sahasrabudhe received a UNC Summer Undergraduate Research Fellowship to use CRISPR technology to study the mechanisms of DNA looped based control of transcription in human cells.

01|23|20
Josh Coon speaking at UNC
Doug will be co-hosting (with Leslie Hicks) Josh Coon's lecture as part of the Analytical Chemistry seminar series:
New Technologies for Global & Rapid Protein, Lipid & Metabolite Measurements
Josh Coon
Professor of Chemistry & Biomolecular Chemistry, University of Wisconsin-Madison
Director, NIGMS National Center for Quanitative Biology of Complex Systems
Feb. 3 — 12:20 pm — Chapman 125
01|09|20
New Alzheimer’s Manuscript published in JoVE
In collaboration with Hyejung Won’s lab here at UNC, we recently published a paper in JOVE describing how to use Hi-C data to interpret GWAS variants. As a test case we applied this approach to identify sets of putative Alzheimer’s associated genes using Hi-C data from human adult brain tissue. More info can be found in the full article.
12|11|19
Team GENE-gerloops wins the TIBBS gingerbread house contest
Our 11-member cookie construction powerhouse took the cake for structural integrity and attention to detail.
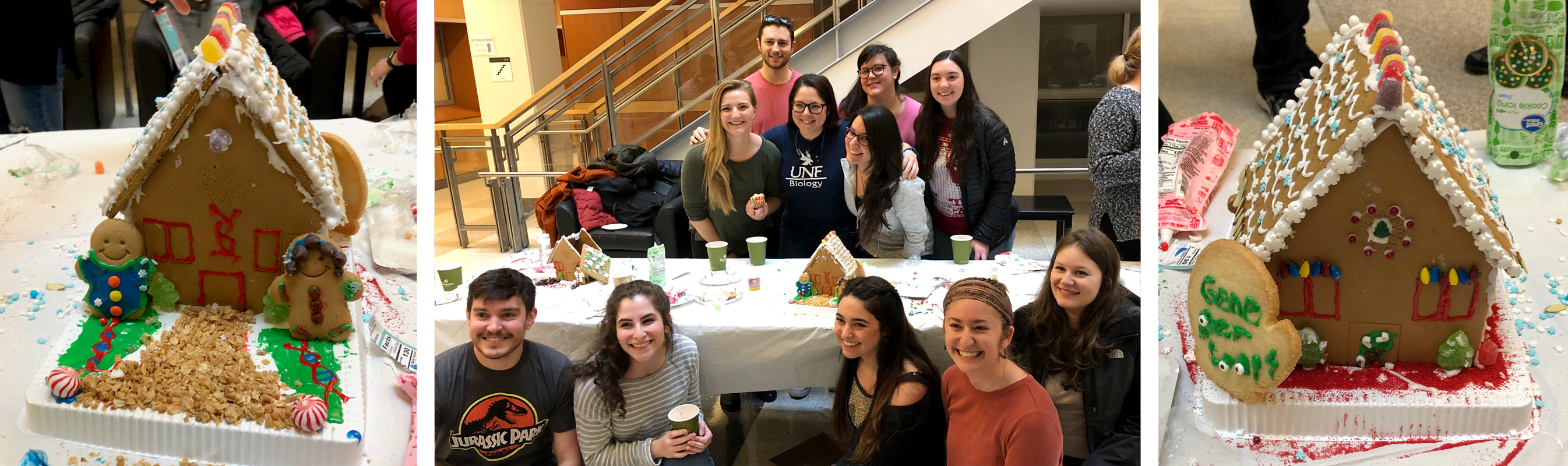
12|06|19
Two papers first-authored by undergrads published in JOSS
Bedtoolsr: An R package for genomic data analysis and manipulation, written by Mayura Patwardhan, and CoralP: Flexible visualization of the human phosphatome, written by Amit Min, were both published this week in The Journal of Open Source Software.
11|12|19
Eliza passes GMB Candidacy Exam
Congratulations to Eliza Thulson for passing the Genetics and Molecular Biology doctoral oral candidacy exam. Now a Ph.D. candidate, Eliza will continue to work on her thesis project studying the mechanisms and disease contributions of DNA looping.

10|25|19
Welcome Agustin and Shannon
Agustin is a post doc who is developing new dCas9 constructs to explore the epigenome. Shannon is a research technician who will be studying how chromatin accessibility changes over biological time courses.
08|20|19
Yoseli and Eric win poster awards at Genetics Retreat
Congratulations to Yoseli and Eric for their poster awards at the UNC Scientific Retreat with the Department of Genetics, the Curriculum in Genetics and Molecular Biology, and the Curriculum in Bioinformatics and Computational Biology. Yoseli presented the identification of novel Alzheimer's risk variants using IPSC-derived microglia; Eric presented Lure, a probe design tool for hybrid capture Hi-C (Hi-C2). Thanks to co-authors Katie, Marielle and Nicole, and to Erika for graphics.

08|20|19
Welcome Jenny
Jenny Giannini joins the lab as a BBSP rotation student after a year as a Fulbright research fellow at the Berlin Institute for Medical Systems Biology. She will be optimizing ATAC-seq protocols for different cell types.
07|14|19
Phosphatase visualization software released
Amit, Erika, Marielle, Eric, and Doug have developed CoralP, a user-friendly web application for visualizing human phosphatase data. Just like Coral, our kinase visualization tool, the highly customizable design allows encoding of multiple qualitative and quantititative attributes, offers three visualization modes, and generates publication-quality vector graphic files. To learn more, read the preprint and watch the video for Coral.
07|02|19
Welcome Eden & Yoli
Eden Cruikshank joins the lab as a PREP Scholar and will be studying how activation by misfolded proteins contributes to neurodegenerative diseases. Yoli Meydan is a computer science and biology major and will be using D3 to visualize various lab metrics.
04|29|19
Welcome Back Nicole
Nicole Kramer, who rotated with us last fall, has officially joined the lab as a graduate student. She will be developing an R package to visualize and plot genomic data sets and contributing to pipelines for data analysis.
04|10|19
Robert receives a Summer Undergraduate Research Fellowship
Congratulations to Robert Fisher, who has received a Summer Undergraduate Research Fellowship (SURF) to study transcriptional regulation in macrophages using CRISPR!
04|09|19
Eliza receives an NSF Graduate Research Fellowship
Congratulations to Eliza Thulson, who was awarded an Graduate Research Fellowship from the National Science Foundation! This prestigious award will fund Eliza for the next three years to study the mechanisms of enhancer based gene regulation in human cells.
04|08|19
Eric and Eliza receive travel grants
Congratulations to Eric Davis and Eliza Thulson who each earned a UNC Graduate Student Transportation Grant to present their research at the Keystone Symposia on "3D Genome: Gene Regulation and Disease" in Banff, Alberta, Canada!

03|13|19
Ann Dean speaking at UNC
Doug will be hosting Ann Dean's lecture as part of the Cell Biology & Physiology seminar series:
Enhancer Function, Chromosome Folding, and Gene Transcription
Ann Dean
Senior Investigator
Gene Regulation & Development Section
Laboratory of Cellular & Developmental Biology
NIDDK–National Institutes of Health
Mar. 25 — 12:00 pm — 124 Taylor Hall
02|11|19
Welcome Yiran & Amit, and Welcome Back Eric
Many exciting new arrivals: We welcome Yiran, who is rotating in the group to develop Hi-C protocols, Amit Min, a new undergraduate researcher who is developing software to visualize human phosphatase data, and Eric Davis, who officially joined the group as a graduate student today and will work on computational approaches to study 3D chromatin architecture. We are thrilled to have them all on board!
02|08|19
Alistair Boettiger & Nils Gehlenborg visit the lab
We hosted two incredible speakers two weeks in a row. Alistair described his work using super-resolution microscopy to study 3D chromatin structures in single cells during development. And Nils demonstrated the theory and power of his lab's HiGlass software for fast and smooth browsing of Hi-C data sets. Both really inspiring talks. It was a great privilege to host them.


01|11|19
Guest Speakers at UNC
Doug will be hosting two lectures next month in the BCB/GMB spring seminar series:
Tracing 3D DNA Structures During Embryogenesis
Alistair Boettiger, Assistant Professor
Stanford Developmental Biology
Feb. 1 — 12:00 pm — Bioinformatics 1131Data Visualization in Biomedical Sciences
Nils Gehlenborg, Assistant Professor
Harvard Biomedical Informatics
Feb. 8 — 12:00 pm — Bioinformatics 1131


11|15|18
Welcome Ennessa
We are happy to welcome BBSP rotation student Ennessa Curry, who will be working on characterizing cell-line-specific genome editing efficiencies.

09|26|18
Coral paper published in Cell Systems
Work from the lab led by Katie Metz and Erika Deoudes was published in the current issue of Cell Systems and featured on the cover. The paper describes Coral, a user-friendly interactive web application for visualizing both quantitative and qualitative data of the human kinome. Coral can encode data in three features (node color, node size, and branch color), allows three modes of kinome visualization (the traditional kinome tree as well as radial and dynamic force networks), and generates high-resolution scalable vector graphics files suitable for publication without the need for refinement using graphics editing software.
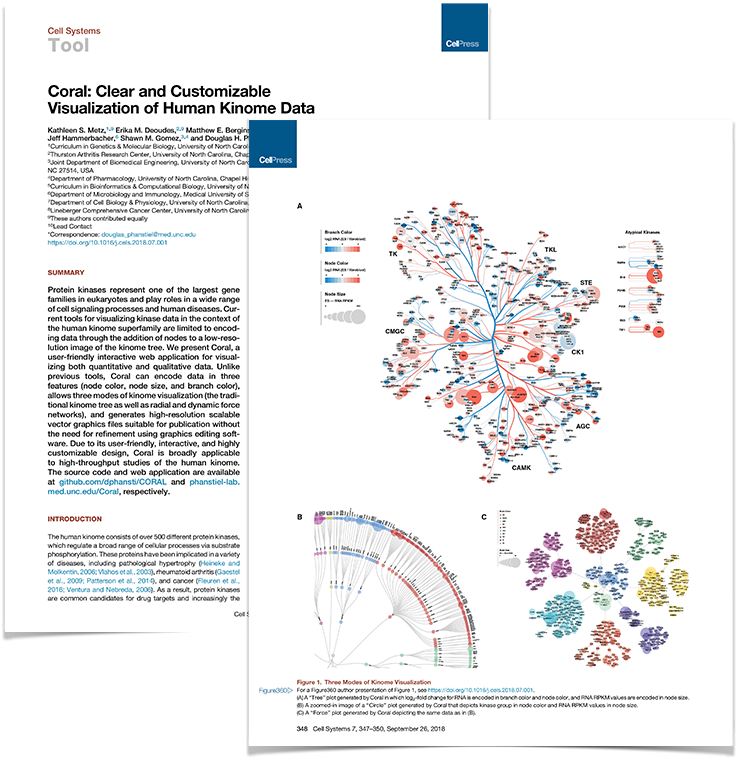
09|01|18
Welcome Isha
We are happy to welcome Isha Sahasrabudhe, a freshman at UNC who will be assisting with CRISPR-based genome editing and general lab continuity.
08|27|18
Welcome Nicole and Caitlin
We are excited to welcome Nicole Kramer and Caitlin Johnson, who are starting rotations in the lab. Nicole hails from New Jersey and will be developing software and wetlab methods for targeted studies of DNA looping. Caitlin joins us from Pennsylvania and is using RNA seq to study mechanisms of osteoarthritis progression.
08|01|18
Welcome Yoseli
Today the lab welcomes our first post doc, Yoseli Quiroga. Yoseli completed her PhD in the Molecular Biology Laboratory of the The Biochemistry Research Institute of La Plata, Argentina. Her work in our lab will focus on understanding how combinatorial transcription factor binding patterns contribute to loop-based gene regulation during cellular differentiation.
07|19|18
The lab receives 5 years of funding from NIGMS
We are grateful to have been awarded a 5-Year, $1.25 million, R35 MIRA (Maximizing Investigators’ Research Award) grant from the National Institute of General Medical Sciences (NIGMS) for research into how human cells regulate gene transcription during development. This research is focused on better understanding how DNA folds within the cell nucleus, specifically which proteins are involved in governing that folding, and how the resulting three-dimensional structure of chromatin regulates gene transcription. The proposed work entails generating new maps of chromatin loops during human development, developing new software to interpret and visualize the resulting data sets, and using genome-editing to identify the mechanisms of cell-type specific looping.

06|22|18
New funding to study tumor-associated macrophage development
During breast cancer progression tumor cells recruit genotypically normal monocytes and drive their differentiation into specialized cells called tumor-associated macrophages (TAMs). TAMs are essential for tumor growth and facilitate multiple tumor promoting activities including vascularization, tissue invasion, and immune suppression making them intriguing targets for therapeutic interventions. We received funding from the Lineberger Cancer Research Center to explore the regulatory mechanisms that drive TAM development and to develop high-throughput genome-editing approaches to inhibit TAM formation.
06|01|18
Eliza receives funding from T32 training grant
Congratulations to Eliza, who earned a one year appointment to an NIGMS T32 Genetics training grant!
06|01|18
Welcome Marielle and Eric
Eric Davis starts his BBSP summer rotation in the group next week. Marielle Bond comes on board as a research technician.
05|25|18
New kinase visualization software released

Katie, Erika, Doug, and several outside contributors have developed Coral, a user-friendly interactive web application for visualizing human kinome data. The highly customizable design allows encoding of multiple qualitative and quantititative attributes, offers three visualization modes, and generates publication-quality vector graphic files. To learn more, read the preprint and watch the video.
05|15|18
Eliza joins the group
We are excited to announce that Eliza Thulson has joined the group as a GMB graduate student. She will be using genome editing to decipher the mechanisms of long-range gene regulatin during macrophage development.
02|11|18
Welcome Eliza and Maya
We are excited to welcome two new lab members this week: Eliza Thulson joins the group as a BBSP rotation student to work on transcription factor-mediated DNA loop formation during macrophage development. Mayura Patwardhan is an undergraduate researcher developing a new R package for genomic data analysis.
01|25|18
Katie receives a travel grant
Congratulations to Katie Metz who was awarded a UNC Graduate Student Transportation Grant to help offset costs for her travel to the a Keystone Symposia. She will be traveling to Whistler, British Columbia, Canada next month to present her research at the 'Chromatin Architecture and Chromosome Organization' meeting.
01|10|18
Welcome Robert
Robert Fisher joins the group as an undergraduate researcher. He will employ CRISPR/Cas9 technology to study the role of AP-1 in enhancer-mediated gene regulation.
12|07|17
Doug receives a JFD Award
Doug has received a Junior Faculty Development Award to develop high-throughput CRISPR screens of chromatin loops involved in macrophage development.