Introducing Lure
A probe design tool for fishing Hi-C data
Lure is a fast, user-friendly web application for designing oligonucleotide probes for hybrid capture Hi-C (Hi-C2). Developed by the Aiden Lab , Hi-C2 allows scientists to enrich Hi-C data in a small, continuous genomic region (Sanborn et al. 2015). Lure is tailored specifically to Hi-C2, designing the optimal probes for Hi-C library enrichment.
Quick Start
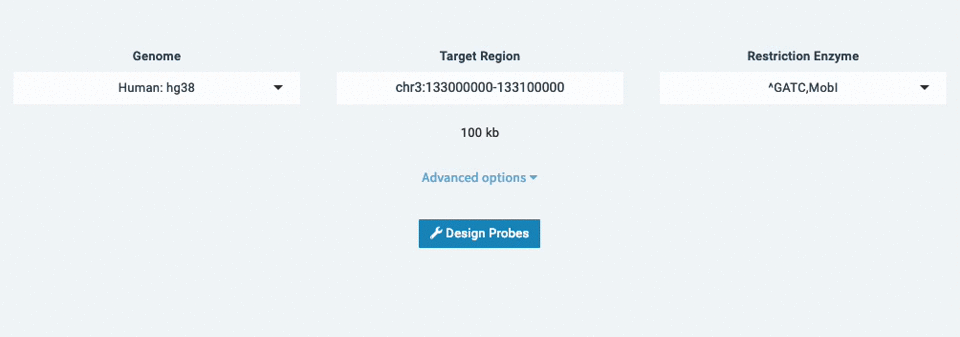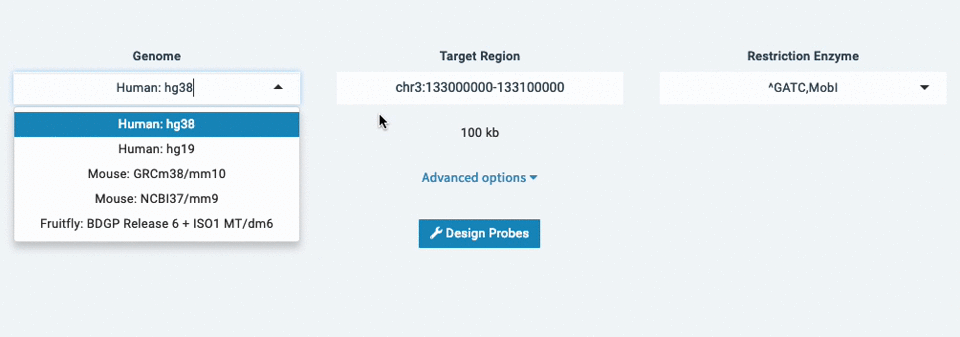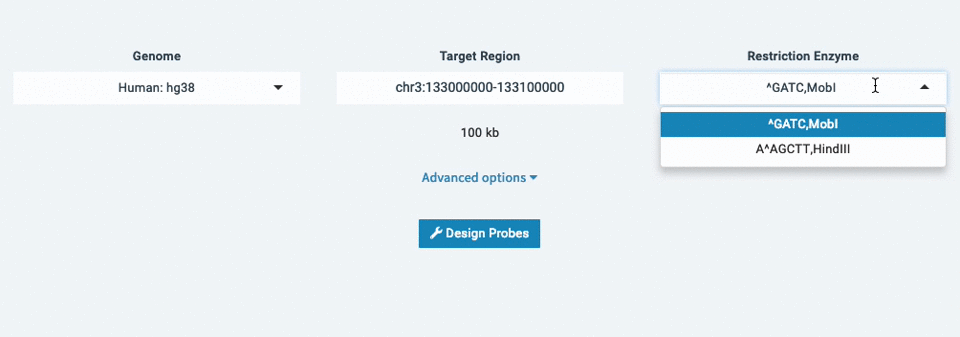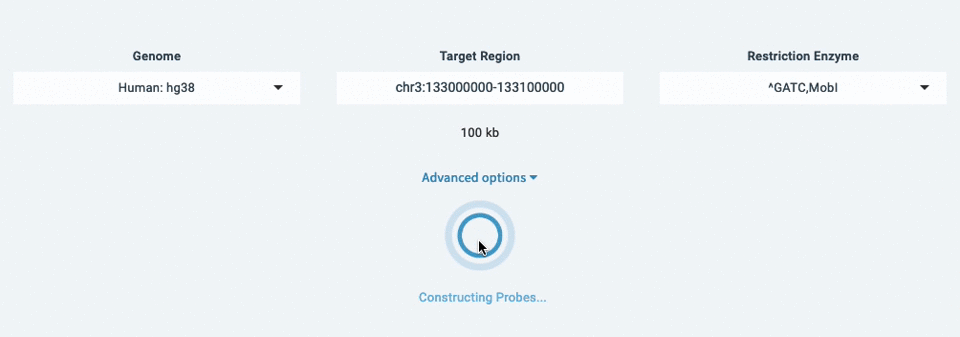
Designing probes with Lure is fast, easy, and doesn't require downloads or dependencies. To get started:
- Select a genome (ex: Human: hg38)
- Enter your target region (ex: chr3:133,000,000-133,100,000)
- Choose your Restriction Enzyme (ex: ^GATC,MobI)
- Design Probes!
How to Cite
Lure was developed in the Phanstiel Lab at UNC by Eric Davis, Erika Deoudes, Craig Wenger, and Doug Phanstiel.
If you use this webapp to design your probes, please cite:
Davis ES, Deoudes EM, Thulson EA, Kramer N, Wenger C and Phanstiel DH. Lure: A Probe Design Tool for Hybrid-Capture Hi-C. In Progress. 2019.
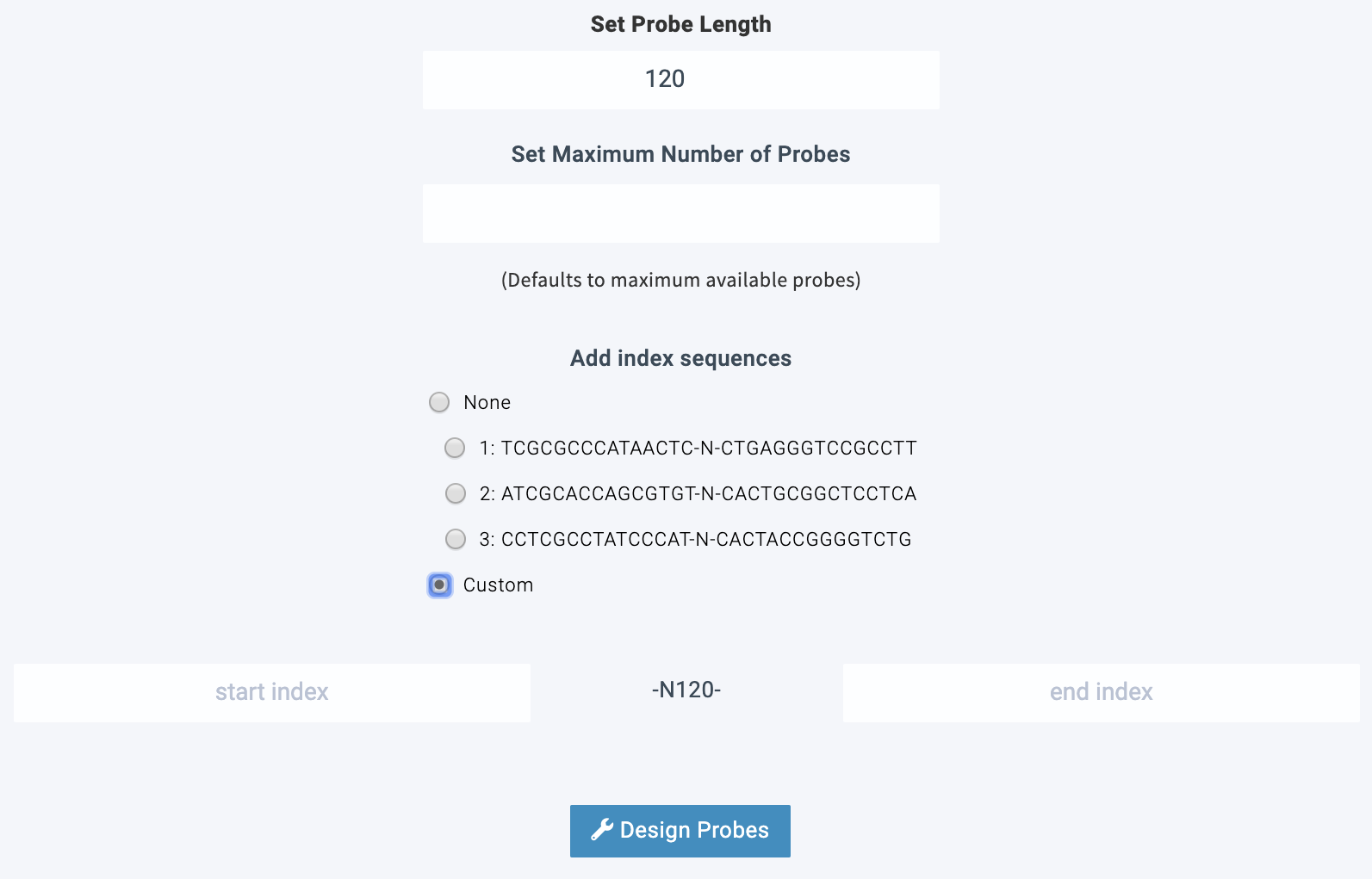
Advanced options
Lure offers advanced options for adjusting the number of desired probes and adding predefined or custom index sequences to the start and end of each probe sequence. This allows for pooling of probes to reduce cost.
How it Works
Implementation
The Lure web application is a Shiny R implementation of the Lure command line tool, which is written primarily using BASH and R. It utilizes several command line tools to identify and fragment the region of interest (ROI) by restriction site (HiCUP - Hi-C User Pipeline), extract genomic fragments (Bedtools), and assess potential probes in parallel (GNU Parallel). The diagram below outlines LURE's workflow:
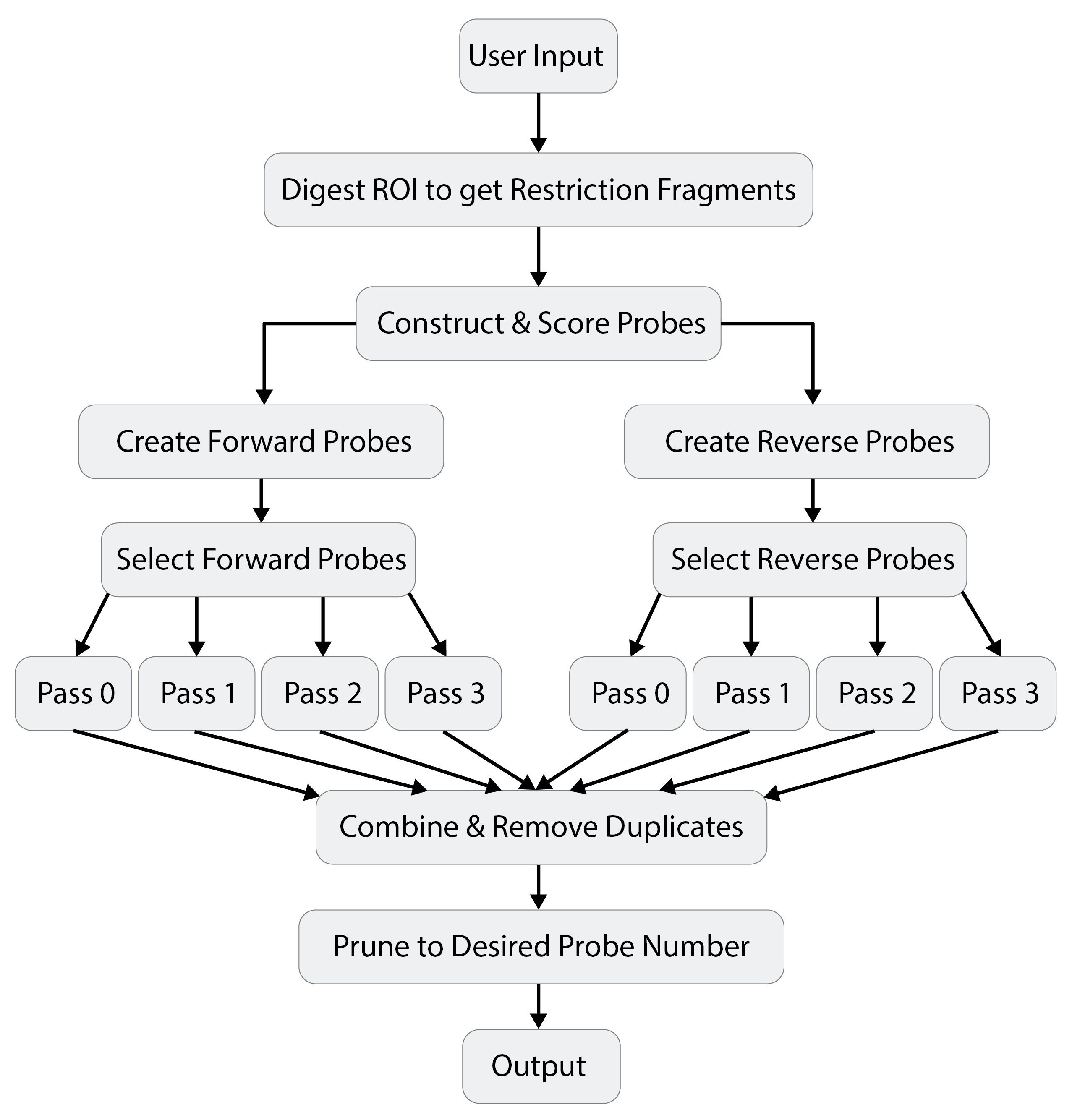
Selection
Criteria for selecting probes were modeled after Sanborn et al. (2015) where they outline their Hi-C2 method. Briefly, potential probes were identified upstream and downstream of each restriction site. All potential probes were scored for repetitive bases, GC content, and distance from restriction site. Each probe was assigned a "Pass Number" (0-3) for overall quality according to the scheme below: